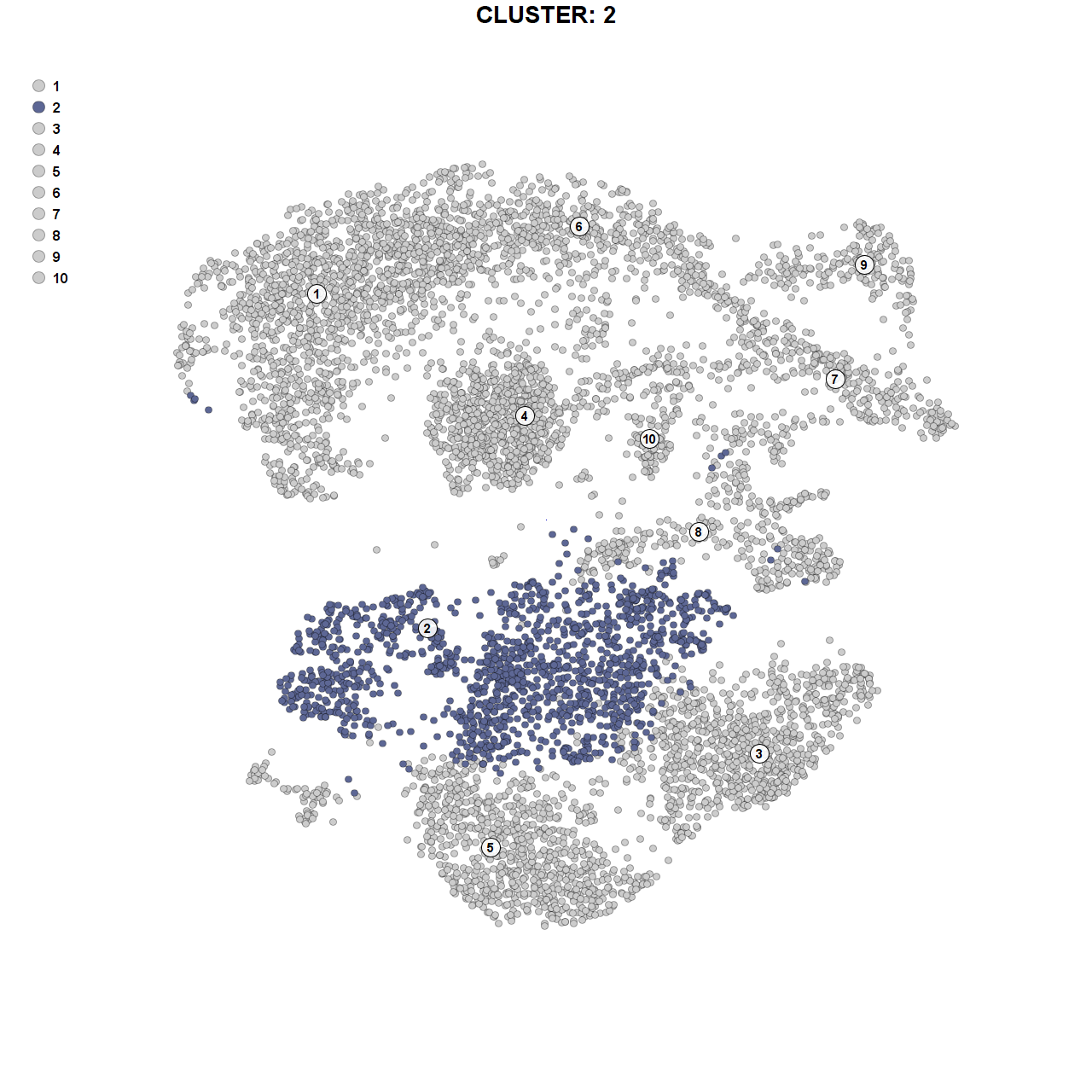
Variable genes for cluster 2
| Nr. | Gene | fc | pv | Synonyms | Ensembl | Details |
|---|---|---|---|---|---|---|
| 1. | GDF15 | 5.549 | 0.000 | GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | ENSG00000130513 | Details |
| 2. | DDIT3 | 4.316 | 0.000 | AltDDIT3, C/EBPzeta, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | ENSG00000175197 | Details |
| 3. | SOD2 | 4.104 | 0.000 | GC1, GClnc1, IPO-B, IPOB, MNSOD, MVCD6, Mn-SOD | ENSG00000291237 | Details |
| 4. | GADD45A | 3.017 | 0.000 | DDIT1, GADD45 | ENSG00000116717 | Details |
| 5. | UPP1 | 2.887 | 0.000 | UDRPASE, UP, UPASE, UPP | ENSG00000183696 | Details |
| 6. | TXNIP | 2.798 | 0.000 | ARRDC6, EST01027, HHCPA78, THIF, VDUP1 | ENSG00000265972 | Details |
| 7. | SQSTM1 | 2.596 | 0.000 | A170, DMRV, FTDALS3, NADGP, OSIL, PDB3, ZIP3, p60, p62, p62B | ENSG00000161011 | Details |
| 8. | RND3 | 2.541 | 0.000 | ARHE, Rho8, RhoE, memB | ENSG00000115963 | Details |
| 9. | STC1 | 2.516 | 0.000 | STC | ENSG00000159167 | Details |
| 10. | MDM2 | 2.395 | 0.000 | ACTFS, HDMX, LSKB, hdm2 | ENSG00000135679 | Details |
| 11. | DCN | 2.352 | 0.000 | CSCD, DSPG2, PG40, PGII, PGS2, SLRR1B | ENSG00000011465 | Details |
| 12. | AKR1B1 | 2.346 | 0.000 | ADR, ALDR1, ALR2, AR | ENSG00000085662 | Details |
| 13. | LUM | 2.199 | 0.000 | LDC, SLRR2D | ENSG00000139329 | Details |
| 14. | LGALS3 | 2.194 | 0.000 | CBP35, GAL3, GALBP, GALIG, L31, LGALS2, MAC2 | ENSG00000131981 | Details |
| 15. | B2M | 2.175 | 0.000 | IMD43 | ENSG00000166710 | Details |
| 16. | HLA-B | 2.174 | 0.000 | AS, B-4901, HLAB | ENSG00000234745 | Details |
| 17. | DDIT4 | 2.119 | 0.000 | Dig2, REDD-1, REDD1 | ENSG00000168209 | Details |
| 18. | MX1 | 2.114 | 0.000 | IFI-78K, IFI78, MX, MxA, lncMX1-215 | ENSG00000157601 | Details |
| 19. | IFIT1 | 2.109 | 0.000 | C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | ENSG00000185745 | Details |
| 20. | ISG15 | 2.070 | 0.000 | G1P2, IFI15, IMD38, IP17, UCRP, hUCRP | ENSG00000187608 | Details |
| 21. | IFITM1 | 2.011 | 0.000 | 9-27, CD225, DSPA2a, IFI17, LEU13 | ENSG00000185885 | Details |
| 22. | IER3 | 2.292 | 2.409e-303 | DIF-2, DIF2, GLY96, IEX-1, IEX-1L, IEX1, PRG1 | ENSG00000137331 | Details |
| 23. | SERPINE2 | 2.160 | 2.488e-302 | GDN, GDNPF, PI-7, PI7, PN-1, PN1, PNI | ENSG00000135919 | Details |
| 24. | GPNMB | 2.060 | 5.259e-297 | HGFIN, NMB, PLCA3 | ENSG00000136235 | Details |
| 25. | PHLDA1 | 2.250 | 6.738e-280 | DT1P1B11, PHRIP, TDAG51 | ENSG00000139289 | Details |
| 26. | PLIN2 | 3.466 | 6.696e-277 | ADFP, ADRP | ENSG00000147872 | Details |
| 27. | SAT1 | 2.985 | 1.020e-271 | DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | ENSG00000130066 | Details |
| 28. | NFKBIA | 2.597 | 1.465e-267 | EDAID2, IKBA, MAD-3, NFKBI | ENSG00000100906 | Details |
| 29. | CTSK | 3.126 | 2.531e-265 | CTS02, CTSO, CTSO1, CTSO2, PKND, PYCD | ENSG00000143387 | Details |
| 30. | OAS1 | 2.451 | 1.757e-262 | E18/E16, IFI-4, IMD100, OIAS, OIASI | ENSG00000089127 | Details |
| 31. | SLC3A2 | 2.417 | 3.395e-254 | 4F2, 4F2HC, 4T2HC, CD98, CD98HC, MDU1, NACAE | ENSG00000168003 | Details |
| 32. | DNAJB9 | 2.037 | 3.159e-235 | ERdj4, MDG-1, MDG1, MST049, MSTP049 | ENSG00000128590 | Details |
| 33. | APOD | 3.768 | 8.911e-220 | - | ENSG00000189058 | Details |
| 34. | BTG2 | 2.318 | 1.217e-218 | APRO1, PC3, TIS21 | ENSG00000159388 | Details |
| 35. | CDKN1A | 2.506 | 3.514e-212 | CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | ENSG00000124762 | Details |
| 36. | BTG1 | 2.218 | 1.818e-211 | APRO2 | ENSG00000133639 | Details |
| 37. | IFIH1 | 2.384 | 4.048e-207 | AGS7, Hlcd, IDDM19, IMD95, MDA-5, MDA5, RLR-2, SGMRT1 | ENSG00000115267 | Details |
| 38. | IFRD1 | 2.247 | 1.511e-204 | PC4, TIS7 | ENSG00000006652 | Details |
| 39. | FBXO32 | 3.812 | 3.648e-202 | Fbx32, MAFbx | ENSG00000156804 | Details |
| 40. | EREG | 2.619 | 2.928e-171 | EPR, ER, Ep | ENSG00000124882 | Details |
| 41. | LURAP1L | 2.139 | 3.928e-170 | C9orf150, HYST0841, LRAP35b, bA3L8.2 | ENSG00000153714 | Details |
| 42. | CXCL2 | 4.527 | 1.209e-161 | CINC-2a, GRO2, GROb, MGSA-b, MIP-2a, MIP2, MIP2A, SCYB2 | ENSG00000081041 | Details |
| 43. | RRAD | 3.648 | 7.098e-150 | RAD, RAD1, REM3 | ENSG00000166592 | Details |
| 44. | IL6 | 3.285 | 1.145e-147 | BSF-2, BSF2, CDF, HGF, HSF, IFN-beta-2, IFNB2, IL-6 | ENSG00000136244 | Details |
| 45. | IL1B | 3.658 | 4.600e-145 | IL-1, IL1-BETA, IL1F2, IL1beta | ENSG00000125538 | Details |
| 46. | ATF3 | 2.765 | 1.166e-144 | - | ENSG00000162772 | Details |
| 47. | KRT17 | 2.546 | 6.600e-144 | 39.1, CK-17, K17, PC, PC2, PCHC1 | ENSG00000128422 | Details |
| 48. | TFPI2 | 2.592 | 8.152e-141 | PP5, REF1, TFPI-2 | ENSG00000105825 | Details |
| 49. | CXCL8 | 7.700 | 5.681e-140 | GCP-1, GCP1, IL8, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1, SCYB8 | ENSG00000169429 | Details |
| 50. | CCN3 | 2.084 | 7.023e-135 | IBP-9, IGFBP-9, IGFBP9, NOV, NOVh | ENSG00000136999 | Details |
| 51. | AVPI1 | 2.008 | 4.856e-133 | PP5395, VIP32, VIT32 | ENSG00000119986 | Details |
| 52. | IRF1 | 2.226 | 4.560e-129 | IRF-1, MAR | ENSG00000125347 | Details |
| 53. | PMAIP1 | 3.483 | 2.326e-126 | APR, NOXA | ENSG00000141682 | Details |
| 54. | ARRDC3 | 2.026 | 1.749e-122 | TLIMP | ENSG00000113369 | Details |
| 55. | HERC5 | 4.567 | 5.796e-121 | CEB1, CEBP1 | ENSG00000138646 | Details |
| 56. | CYP1B1 | 2.382 | 9.351e-120 | ASGD6, CP1B, CYPIB1, GLC3A, P4501B1 | ENSG00000138061 | Details |
| 57. | IFIT2 | 4.502 | 7.224e-112 | G10P2, GARG-39, IFI-54, IFI-54K, IFI54, IFIT-2, ISG-54, ISG-54 K, ISG-54K, ISG54, P54, cig42 | ENSG00000119922 | Details |
| 58. | CLEC2B | 2.518 | 6.880e-111 | AICL, CLECSF2, HP10085, IFNRG1 | ENSG00000110852 | Details |
| 59. | C15orf48 | 2.001 | 7.297e-109 | COXFA4L3, FOAP-11, MIR147BHG, MISTRAV, MOCCI, NMES1 | ENSG00000166920 | Details |
| 60. | HSPA5 | 2.380 | 1.576e-91 | BIP, GRP78, HEL-S-89n | ENSG00000044574 | Details |
| 61. | CLCA2 | 3.055 | 5.268e-90 | CACC, CACC3, CLCRG2, CaCC-3 | ENSG00000137975 | Details |
| 62. | TRIB3 | 2.061 | 5.322e-90 | C20orf97, NIPK, SINK, SKIP3, TRB3 | ENSG00000101255 | Details |
| 63. | AREG | 4.318 | 5.216e-88 | AR, AREGB, CRDGF, SDGF | ENSG00000109321 | Details |
| 64. | SNHG12 | 2.736 | 1.718e-86 | ASLNC04080, C1orf79, LINC00100, NCRNA00100, PNAS-123 | ENSG00000197989 | Details |
| 65. | PLAAT4 | 2.222 | 2.835e-85 | HRASLS4, HRSL4, PLA1/2-3, PLAAT-4, RARRES3, RIG1, TIG3 | ENSG00000133321 | Details |
| 66. | BEX2 | 4.124 | 5.715e-84 | BEX1, DJ79P11.1 | ENSG00000133134 | Details |
| 67. | FABP3 | 2.401 | 6.829e-81 | FABP11, H-FABP, M-FABP, MDGI, O-FABP | ENSG00000121769 | Details |
| 68. | MMP1 | 2.136 | 1.433e-78 | CLG, CLGN | ENSG00000196611 | Details |
| 69. | RBP4 | 3.052 | 2.319e-78 | MCOPCB10, RDCCAS | ENSG00000138207 | Details |
| 70. | MT1H | 7.152 | 2.022e-75 | MT-0, MT-1H, MT-IH, MT1 | ENSG00000205358 | Details |
| 71. | LINC01705 | 3.880 | 1.610e-74 | ERLR | Details | |
| 72. | BIRC3 | 2.077 | 1.499e-73 | AIP1, API2, CIAP2, HAIP1, HIAP1, IAP-1, MALT2, MIHC, RNF49, c-IAP2 | ENSG00000023445 | Details |
| 73. | RSAD2 | 4.785 | 1.152e-72 | SAND, cig33, cig5, vig1 | ENSG00000134321 | Details |
| 74. | CCDC30 | 4.687 | 4.992e-69 | PFD6L, PFDN6L | ENSG00000186409 | Details |
| 75. | CCL5 | 5.435 | 2.873e-67 | D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | ENSG00000271503 | Details |
| 76. | CHI3L2 | 4.333 | 2.671e-65 | CHIL2, YKL-39, YKL39 | ENSG00000064886 | Details |
| 77. | LINC02154 | 2.336 | 5.902e-62 | MALR | ENSG00000235385 | Details |
| 78. | LOC101929048 | 5.459 | 2.869e-61 | - | Details | |
| 79. | SPINK1 | 5.178 | 3.942e-60 | PCTT, PSTI, Spink3, TATI, TCP | ENSG00000164266 | Details |
| 80. | CSF3 | 4.201 | 7.389e-60 | C17orf33, CSF3OS, GCSF | ENSG00000108342 | Details |
| 81. | CXCL1 | 5.268 | 2.201e-59 | FSP, GRO1, GROa, MGSA, MGSA-a, NAP-3, SCYB1 | ENSG00000163739 | Details |
| 82. | LOC284344 | 4.021 | 4.224e-59 | - | ENSG00000241104 | Details |
| 83. | AKR1C1 | 2.187 | 5.578e-59 | 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DD1/DD2, DDH, DDH1, H-37, HAKRC, HBAB, MBAB | ENSG00000187134 | Details |
| 84. | CSF2 | 2.761 | 6.178e-57 | CSF, GMCSF | ENSG00000164400 | Details |
| 85. | ANGPTL2 | 2.053 | 4.138e-56 | ARP2, HARP | ENSG00000136859 | Details |
| 86. | OASL | 4.297 | 2.275e-54 | OASL1, OASLd, TRIP-14, TRIP14, p59 OASL, p59-OASL, p59OASL | ENSG00000135114 | Details |
| 87. | LINC00520 | 2.365 | 2.420e-51 | C14orf34, LASSIE, LEENE, LncRNA00520 | ENSG00000258791 | Details |
| 88. | IFI30 | 2.896 | 1.018e-50 | GILT, IFI-30, IP-30, IP30 | ENSG00000216490 | Details |
| 89. | TNFAIP6 | 4.153 | 2.287e-50 | TSG-6, TSG6 | ENSG00000123610 | Details |
| 90. | ISG20 | 2.958 | 5.745e-49 | CD25, HEM45 | ENSG00000172183 | Details |
| 91. | CCL7 | 2.552 | 7.892e-47 | FIC, MARC, MCP-3, MCP3, NC28, SCYA6, SCYA7 | ENSG00000108688 | Details |
| 92. | LOC107986820 | 4.828 | 1.067e-46 | Details | ||
| 93. | CXCL3 | 4.635 | 1.846e-42 | CINC-2b, GRO3, GROg, MIP-2b, MIP2B, SCYB3 | ENSG00000163734 | Details |
| 94. | FAM20A | 2.090 | 6.940e-41 | AI1G, AIGFS, FP2747 | ENSG00000108950 | Details |
| 95. | C11orf96 | 3.904 | 4.385e-38 | AG2 | ENSG00000187479 | Details |
| 96. | MT1G | 9.623 | 1.738e-33 | MT1, MT1K | ENSG00000125144 | Details |
| 97. | G0S2 | 2.437 | 1.272e-30 | - | ENSG00000123689 | Details |
| 98. | GADD45B | 2.139 | 3.916e-26 | GADD45BETA, MYD118 | ENSG00000099860 | Details |
| 99. | SOX4 | 3.235 | 5.100e-21 | CSS10, EVI16 | ENSG00000124766 | Details |
| 100. | GEM | 2.050 | 8.781e-20 | KIR | ENSG00000164949 | Details |
| 101. | NR1D1 | 2.179 | 2.653e-16 | EAR1, REVERBA, REVERBalpha, THRA1, THRAL, ear-1, hRev | ENSG00000126368 | Details |
| 102. | SAA1 | 5.093 | 3.491e-14 | PIG4, SAA, SAA2, TP53I4 | ENSG00000173432 | Details |
| 103. | MT1X | 2.638 | 6.047e-11 | MT-1l, MT1 | ENSG00000187193 | Details |
Berezikov Lab - 2023 © ERIBA