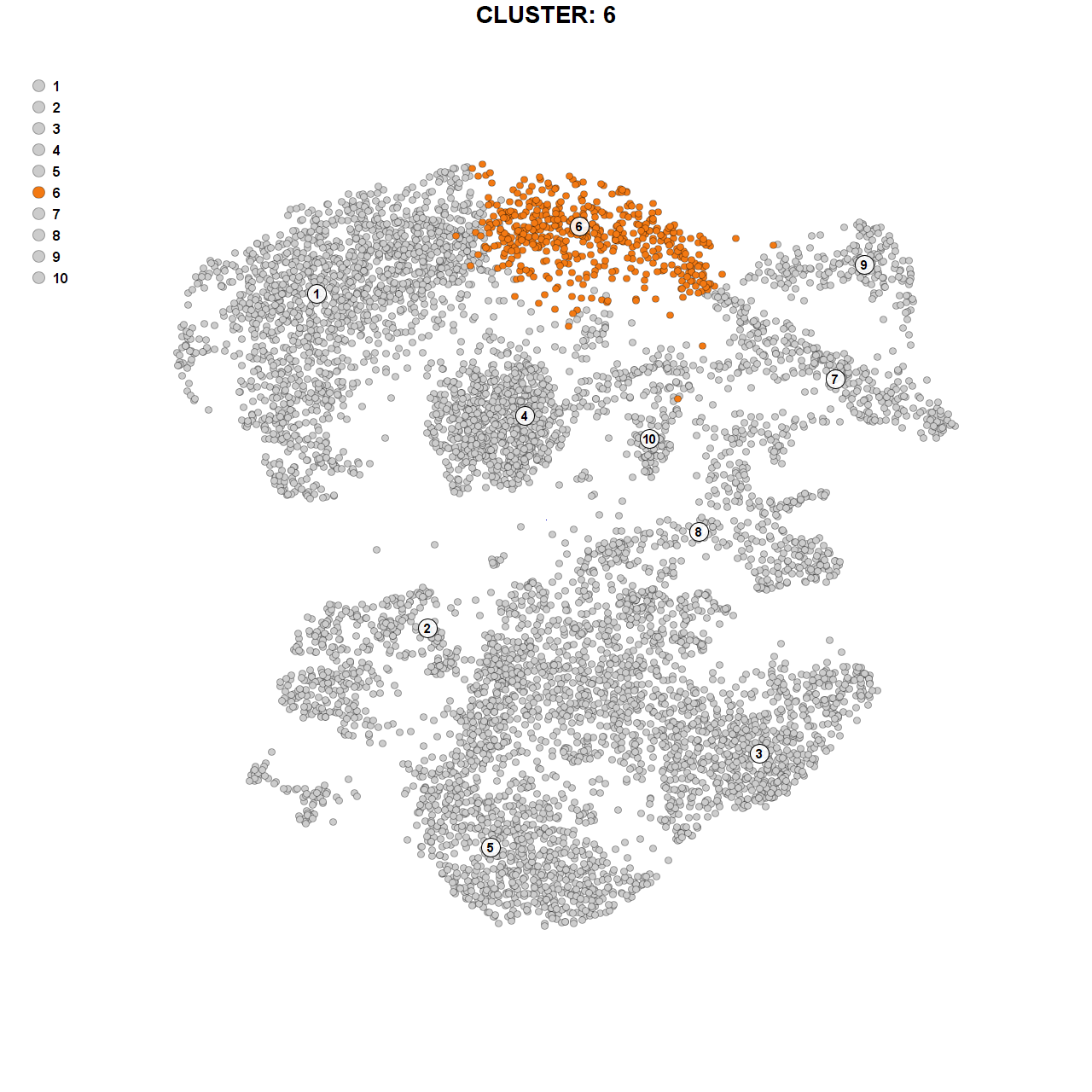
Marker genes for cluster 6
| Nr. | Gene | mean.ncl | mean.cl | fc | pv | padj | Synonyms | Ensembl | Details |
|---|---|---|---|---|---|---|---|---|---|
| 1. | TYMS | 0.034 | 0.083 | 2.417 | 5.817e-06 | 0.000 | DKCD, HST422, TMS, TS | ENSG00000176890 | Details |
| 2. | CDKN3 | 0.036 | 0.085 | 2.368 | 5.834e-06 | 0.000 | CDI1, CIP2, KAP, KAP1 | ENSG00000100526 | Details |
| 3. | PCLAF | 0.036 | 0.084 | 2.344 | 5.454e-06 | 0.000 | KIAA0101, L5, NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | ENSG00000166803 | Details |
| 4. | FUS | 0.047 | 0.098 | 2.104 | 1.025e-05 | 0.000 | ALS6, ETM4, FUS1, HNRNPP2, POMP75, TLS, altFUS | ENSG00000089280 | Details |
| 5. | EBNA1BP2 | 0.051 | 0.104 | 2.048 | 1.020e-05 | 0.000 | EBP2, NOBP, P40 | ENSG00000117395 | Details |
| 6. | TUBA1B | 0.216 | 0.571 | 2.638 | 1.420e-39 | 3.455e-37 | K-ALPHA-1 | ENSG00000123416 | Details |
| 7. | STMN1 | 0.162 | 0.424 | 2.617 | 1.427e-29 | 2.315e-27 | C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | ENSG00000117632 | Details |
| 8. | ID1 | 0.206 | 0.432 | 2.092 | 1.639e-19 | 1.932e-17 | ID, bHLHb24 | ENSG00000125968 | Details |
| 9. | HNRNPA2B1 | 0.193 | 0.407 | 2.106 | 1.126e-18 | 1.252e-16 | HNRNPA2, HNRNPB1, HNRPA2, HNRPA2B1, HNRPB1, IBMPFD2, OPMD2, RNPA2, SNRPB1 | ENSG00000122566 | Details |
| 10. | NQO1 | 0.147 | 0.324 | 2.211 | 1.765e-16 | 1.675e-14 | DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | ENSG00000181019 | Details |
| 11. | OXTR | 0.178 | 0.359 | 2.017 | 3.608e-15 | 2.987e-13 | OT-R, OTR | ENSG00000180914 | Details |
| 12. | PTMA | 0.152 | 0.319 | 2.103 | 6.199e-15 | 5.026e-13 | TMSA | ENSG00000187514 | Details |
| 13. | H2AZ2 | 0.095 | 0.221 | 2.322 | 5.220e-13 | 3.386e-11 | H2A.Z-2, H2AFV, H2AV | ENSG00000105968 | Details |
| 14. | HNRNPD | 0.092 | 0.213 | 2.326 | 6.653e-13 | 4.245e-11 | AUF1, AUF1A, HNRPD, P37, hnRNPD0 | ENSG00000138668 | Details |
| 15. | PTTG1 | 0.049 | 0.142 | 2.865 | 4.652e-12 | 2.624e-10 | EAP1, ECRAR, HPTTG, PTTG, TUTR1 | ENSG00000164611 | Details |
| 16. | JPT1 | 0.098 | 0.203 | 2.065 | 8.504e-10 | 3.940e-08 | ARM2, HN1, HN1A | ENSG00000189159 | Details |
| 17. | CXCL12 | 0.023 | 0.080 | 3.526 | 9.713e-10 | 4.447e-08 | IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1 | ENSG00000107562 | Details |
| 18. | TK1 | 0.030 | 0.092 | 3.082 | 5.080e-09 | 2.126e-07 | TK2 | ENSG00000167900 | Details |
| 19. | C1QBP | 0.092 | 0.189 | 2.059 | 5.492e-09 | 2.274e-07 | COXPD33, GC1QBP, HABP1, SF2AP32, SF2p32, gC1Q-R, gC1qR, p32 | ENSG00000108561 | Details |
| 20. | BIRC5 | 0.037 | 0.104 | 2.782 | 7.491e-09 | 2.945e-07 | API4, EPR-1 | ENSG00000089685 | Details |
| 21. | CENPW | 0.031 | 0.088 | 2.801 | 7.201e-08 | 2.614e-06 | C6orf173, CENP-W, CUG2 | ENSG00000203760 | Details |
| 22. | HMGB1 | 0.059 | 0.131 | 2.215 | 8.702e-08 | 3.107e-06 | HMG-1, HMG1, HMG3, SBP-1 | ENSG00000189403 | Details |
| 23. | LY6K | 0.034 | 0.089 | 2.637 | 1.042e-07 | 3.653e-06 | CT97, HSJ001348, URLC10, ly-6K | ENSG00000160886 | Details |
| 24. | PRKDC | 0.067 | 0.143 | 2.142 | 1.102e-07 | 3.828e-06 | DNA-PKC, DNA-PKcs, DNAPK, DNAPKc, DNPK1, HYRC, HYRC1, IMD26, XRCC7, p350 | ENSG00000253729 | Details |
| 25. | HNRNPR | 0.063 | 0.136 | 2.162 | 1.301e-07 | 4.402e-06 | HNRPR, NEDDFSB, hnRNP-R | ENSG00000125944 | Details |
| 26. | SFPQ | 0.072 | 0.149 | 2.067 | 2.024e-07 | 6.511e-06 | POMP100, PPP1R140, PSF | ENSG00000116560 | Details |
| 27. | HNRNPH3 | 0.052 | 0.117 | 2.226 | 3.905e-07 | 1.126e-05 | 2H9, HNRPH3 | ENSG00000096746 | Details |
| 28. | CCT2 | 0.067 | 0.138 | 2.070 | 7.525e-07 | 2.034e-05 | 99D8.1, CCT-beta, CCTB, HEL-S-100n, PRO1633, TCP-1-beta | ENSG00000166226 | Details |
| 29. | MRPL3 | 0.039 | 0.092 | 2.394 | 1.223e-06 | 3.195e-05 | COXPD9, MRL3, RPML3, uL3m | ENSG00000114686 | Details |
| 30. | CCT6A | 0.061 | 0.126 | 2.050 | 1.756e-06 | 4.410e-05 | CCT-zeta, CCT-zeta-1, CCT6, Cctz, HTR3, MoDP-2, TCP-1-zeta, TCP20, TCPZ, TTCP20 | ENSG00000146731 | Details |
| 31. | ALYREF | 0.029 | 0.077 | 2.615 | 2.263e-06 | 5.538e-05 | ALY, ALY/REF, BEF, REF, THOC4 | ENSG00000183684 | Details |
| 32. | HNRNPM | 0.049 | 0.108 | 2.196 | 2.417e-06 | 5.878e-05 | CEAR, HNRNPM4, HNRPM, HNRPM4, HTGR1, NAGR1, hnRNP M | ENSG00000099783 | Details |
| 33. | SRSF7 | 0.058 | 0.120 | 2.060 | 3.827e-06 | 8.865e-05 | 9G8, AAG3, SFRS7 | ENSG00000115875 | Details |
| 34. | HSPD1 | 0.047 | 0.102 | 2.181 | 3.861e-06 | 8.892e-05 | CPN60, GROEL, HLD4, HSP-60, HSP60, HSP65, HuCHA60, SPG13 | ENSG00000144381 | Details |
| 35. | RPA3 | 0.032 | 0.073 | 2.295 | 2.833e-05 | 0.001 | REPA3, RP-A p14 | ENSG00000106399 | Details |
| 36. | ARHGAP18 | 0.047 | 0.097 | 2.072 | 2.841e-05 | 0.001 | MacGAP, SENEX, bA307O14.2 | ENSG00000146376 | Details |
| 37. | RANBP1 | 0.047 | 0.097 | 2.054 | 2.957e-05 | 0.001 | HTF9A | ENSG00000099901 | Details |
| 38. | SNRPA | 0.019 | 0.050 | 2.597 | 0.000 | 0.002 | Mud1, U1-A, U1A | ENSG00000077312 | Details |
| 39. | DTYMK | 0.024 | 0.056 | 2.336 | 0.000 | 0.003 | CDC8, CONPM, PP3731, TMPK, TYMK | ENSG00000168393 | Details |
| 40. | NASP | 0.027 | 0.060 | 2.255 | 0.000 | 0.003 | FLB7527, HMDRA1, PRO1999 | ENSG00000132780 | Details |
| 41. | SSRP1 | 0.030 | 0.066 | 2.216 | 0.000 | 0.003 | FACT, FACT80, T160 | ENSG00000149136 | Details |
| 42. | PRMT1 | 0.037 | 0.076 | 2.068 | 0.000 | 0.003 | ANM1, HCP1, HRMT1L2, IR1B4 | ENSG00000126457 | Details |
| 43. | MYO10 | 0.041 | 0.082 | 2.007 | 0.000 | 0.003 | MyoX | ENSG00000145555 | Details |
| 44. | GGH | 0.017 | 0.044 | 2.613 | 0.000 | 0.004 | GATD10, GH | ENSG00000137563 | Details |
| 45. | PHF19 | 0.018 | 0.047 | 2.595 | 0.000 | 0.004 | MTF2L1, PCL3, TDRD19B | ENSG00000119403 | Details |
| 46. | MRPL12 | 0.028 | 0.059 | 2.086 | 0.000 | 0.005 | 5c5-2, L12mt, MRP-L31/34, MRPL7, MRPL7/L12, RPML12, bL12m | ENSG00000262814 | Details |
| 47. | NEDD1 | 0.016 | 0.042 | 2.602 | 0.000 | 0.007 | GCP-WD, TUBGCP7 | ENSG00000139350 | Details |
| 48. | FBL | 0.033 | 0.067 | 2.033 | 0.001 | 0.007 | FIB, FLRN, Nop1, RNU3IP1 | ENSG00000105202 | Details |
| 49. | NUDT1 | 0.023 | 0.050 | 2.228 | 0.001 | 0.009 | MTH1 | ENSG00000106268 | Details |
| 50. | GPATCH4 | 0.027 | 0.055 | 2.064 | 0.001 | 0.009 | GPATC4 | ENSG00000160818 | Details |
Berezikov Lab - 2023 © ERIBA