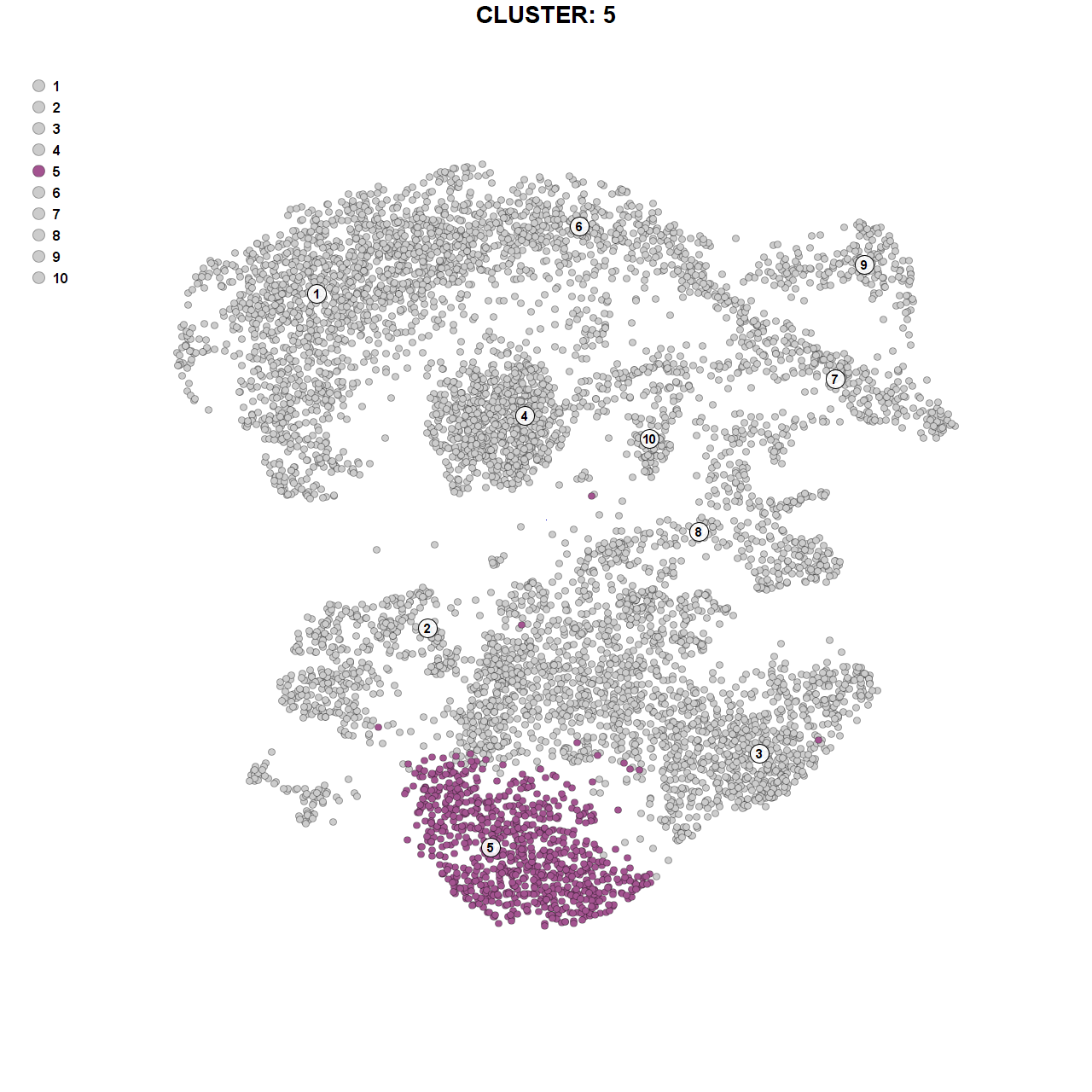
Marker genes for cluster 5
| Nr. | Gene | mean.ncl | mean.cl | fc | pv | padj | Synonyms | Ensembl | Details |
|---|---|---|---|---|---|---|---|---|---|
| 1. | HIF1A-AS3 | 0.004 | 0.017 | 4.706 | 6.212e-05 | 0.000 | HIFAL | ENSG00000258667 | Details |
| 2. | C14orf132 | 0.013 | 0.037 | 2.809 | 2.069e-05 | 0.000 | C14orf88 | ENSG00000227051 | Details |
| 3. | IRS2 | 0.015 | 0.038 | 2.628 | 2.763e-05 | 0.000 | IRS-2 | ENSG00000185950 | Details |
| 4. | IGFBP3 | 1.156 | 2.736 | 2.366 | 0.000 | 0.000 | BP-53, IBP3 | ENSG00000146674 | Details |
| 5. | mito-MT-CYB | 1.686 | 3.687 | 2.187 | 0.000 | 0.000 | Details | ||
| 6. | TRIM56 | 0.026 | 0.055 | 2.070 | 5.195e-05 | 0.000 | RNF109 | ENSG00000169871 | Details |
| 7. | NCOA7 | 0.028 | 0.058 | 2.043 | 5.919e-05 | 0.000 | ERAP140, ESNA1, NCOA7-AS, Nbla00052, Nbla10993, TLDC4, dJ187J11.3 | ENSG00000111912 | Details |
| 8. | mito-MT-CO1 | 3.850 | 7.762 | 2.016 | 0.000 | 0.000 | Details | ||
| 9. | RPL34 | 0.842 | 1.710 | 2.031 | 4.963e-101 | 1.073e-98 | L34, eL34 | ENSG00000109475 | Details |
| 10. | GAS5 | 0.314 | 0.868 | 2.767 | 2.727e-95 | 5.055e-93 | NCRNA00030, SNHG2 | ENSG00000234741 | Details |
| 11. | SNHG6 | 0.248 | 0.723 | 2.911 | 4.144e-86 | 6.204e-84 | HBII-276HG, NCRNA00058, U87HG | Details | |
| 12. | TOMM7 | 0.392 | 0.837 | 2.136 | 1.259e-56 | 1.441e-54 | GMPGS, TOM7 | ENSG00000196683 | Details |
| 13. | RPL12 | 0.396 | 0.808 | 2.040 | 3.611e-49 | 3.603e-47 | L12, uL11 | ENSG00000197958 | Details |
| 14. | BRI3 | 0.357 | 0.729 | 2.040 | 8.031e-45 | 7.269e-43 | I3 | ENSG00000164713 | Details |
| 15. | ZFAS1 | 0.315 | 0.659 | 2.092 | 4.137e-43 | 3.578e-41 | C20orf199, HSUP1, HSUP2, NCRNA00275, ZNFX1-AS1 | ENSG00000177410 | Details |
| 16. | SNHG32 | 0.189 | 0.438 | 2.323 | 2.602e-36 | 1.875e-34 | C6orf48, D6S57, G8 | ENSG00000204387 | Details |
| 17. | SNHG8 | 0.161 | 0.390 | 2.422 | 6.594e-35 | 4.502e-33 | LINC00060, NCRNA00060 | ENSG00000269893 | Details |
| 18. | MMP2 | 0.142 | 0.307 | 2.159 | 2.514e-22 | 1.052e-20 | CLG4, CLG4A, MMP-2, MMP-II, MONA, TBE-1 | ENSG00000087245 | Details |
| 19. | CTSK | 0.077 | 0.194 | 2.533 | 9.839e-20 | 3.829e-18 | CTS02, CTSO, CTSO1, CTSO2, PKND, PYCD | ENSG00000143387 | Details |
| 20. | SCN9A | 0.048 | 0.145 | 3.015 | 1.803e-19 | 6.746e-18 | ETHA, FEB3B, GEFSP7, HSAN2D, NE-NA, NENA, Nav1.7, PN1, SFNP | ENSG00000169432 | Details |
| 21. | STC2 | 0.081 | 0.197 | 2.429 | 1.458e-18 | 5.066e-17 | STC-2, STCRP | ENSG00000113739 | Details |
| 22. | NPC2 | 0.116 | 0.249 | 2.145 | 3.913e-18 | 1.302e-16 | EDDM1, HE1 | ENSG00000119655 | Details |
| 23. | SNHG7 | 0.044 | 0.131 | 2.970 | 2.279e-17 | 7.330e-16 | NCRNA00061 | ENSG00000233016 | Details |
| 24. | MEG3 | 0.097 | 0.216 | 2.234 | 2.358e-17 | 7.522e-16 | FP504, GTL2, LINC00023, NCRNA00023, PRO0518, PRO2160, onco-lncRNA-83, prebp1 | ENSG00000214548 | Details |
| 25. | SLC5A3 | 0.022 | 0.087 | 3.888 | 2.186e-16 | 6.647e-15 | BCW2, SMIT, SMIT1, SMIT2 | ENSG00000198743 | Details |
| 26. | VEGFB | 0.051 | 0.131 | 2.593 | 2.638e-14 | 6.983e-13 | VEGFL, VRF | ENSG00000173511 | Details |
| 27. | H2BC12 | 0.084 | 0.183 | 2.171 | 4.448e-14 | 1.170e-12 | H2B/S, H2BFAiii, H2BFT, H2BK, HIST1H2BK | ENSG00000197903 | Details |
| 28. | SMIM3 | 0.035 | 0.102 | 2.919 | 1.178e-13 | 2.963e-12 | C5orf62, MST150, NID67 | ENSG00000256235 | Details |
| 29. | EPB41L4A-AS1 | 0.048 | 0.120 | 2.496 | 3.959e-12 | 8.856e-11 | C5orf26, NCRNA00219, TIGA1 | Details | |
| 30. | MMP14 | 0.044 | 0.110 | 2.525 | 7.554e-12 | 1.652e-10 | MMP-14, MMP-X1, MT-MMP, MT-MMP 1, MT1-MMP, MT1MMP, MTMMP1, WNCHRS | ENSG00000157227 | Details |
| 31. | ZMAT3 | 0.038 | 0.099 | 2.637 | 2.317e-11 | 4.797e-10 | PAG608, WIG-1, WIG1 | ENSG00000172667 | Details |
| 32. | SEMA5A | 0.036 | 0.095 | 2.645 | 2.557e-11 | 5.237e-10 | SEMAF, semF | ENSG00000112902 | Details |
| 33. | BEST1 | 0.060 | 0.133 | 2.228 | 3.150e-11 | 6.353e-10 | ARB, BEST, BMD, Best1V1Delta2, RP50, TU15B, VMD2 | ENSG00000167995 | Details |
| 34. | SMIM29 | 0.030 | 0.085 | 2.857 | 4.531e-11 | 8.997e-10 | C6orf1, LBH | ENSG00000186577 | Details |
| 35. | EPAS1 | 0.047 | 0.111 | 2.365 | 1.932e-10 | 3.609e-09 | ECYT4, HIF2A, HLF, MOP2, PASD2, bHLHe73 | ENSG00000116016 | Details |
| 36. | LOC124902453 | 0.015 | 0.053 | 3.405 | 1.787e-09 | 2.985e-08 | - | Details | |
| 37. | MT1L | 0.058 | 0.122 | 2.103 | 3.168e-09 | 5.203e-08 | MT1, MT1R, MTF | ENSG00000291105 | Details |
| 38. | CHPF | 0.048 | 0.106 | 2.200 | 3.940e-09 | 6.337e-08 | CHPF1, CHSY2, CSS2 | ENSG00000123989 | Details |
| 39. | GRINA | 0.066 | 0.132 | 2.022 | 4.362e-09 | 6.929e-08 | HNRGW, LFG1, NMDARA1, TMBIM3 | ENSG00000178719 | Details |
| 40. | THBS2 | 0.057 | 0.119 | 2.082 | 5.807e-09 | 9.040e-08 | TSP2 | ENSG00000186340 | Details |
| 41. | PAM | 0.057 | 0.116 | 2.047 | 2.905e-08 | 4.126e-07 | PAL, PAM-1, PHM | ENSG00000145730 | Details |
| 42. | ARL4C | 0.006 | 0.030 | 4.973 | 3.711e-08 | 5.177e-07 | ARL7, LAK | ENSG00000188042 | Details |
| 43. | PIEZO1 | 0.030 | 0.073 | 2.410 | 9.753e-08 | 1.287e-06 | DHS, ER, FAM38A, LMPH3, LMPHM6, Mib | ENSG00000103335 | Details |
| 44. | SCD | 0.028 | 0.069 | 2.483 | 1.060e-07 | 1.394e-06 | FADS5, MSTP008, SCD1, SCDOS, hSCD1 | ENSG00000099194 | Details |
| 45. | PAPPA | 0.047 | 0.097 | 2.074 | 1.515e-07 | 1.918e-06 | ASBABP2, DIPLA1, IGFBP-4ase, PAPA, PAPP-A, PAPPA1 | ENSG00000182752 | Details |
| 46. | INHBB | 0.005 | 0.025 | 5.257 | 4.225e-07 | 5.034e-06 | - | ENSG00000163083 | Details |
| 47. | CELF2 | 0.008 | 0.031 | 3.984 | 4.379e-07 | 5.196e-06 | BRUNOL3, CELF-2, CUG-BP2, CUGBP2, DEE97, ETR-3, ETR3, NAPOR | ENSG00000048740 | Details |
| 48. | IGF2R | 0.025 | 0.061 | 2.440 | 7.278e-07 | 8.306e-06 | CD222, CI-M6PR, CIMPR, M6P-R, M6P/IGF2R, MPR 300, MPR1, MPR300, MPRI | ENSG00000197081 | Details |
| 49. | FAM43A | 0.020 | 0.051 | 2.599 | 9.089e-07 | 1.014e-05 | - | ENSG00000185112 | Details |
| 50. | RPL34P18 | 0.027 | 0.064 | 2.357 | 1.236e-06 | 1.355e-05 | RPL34_7_926 | ENSG00000240509 | Details |
| 51. | VAT1 | 0.042 | 0.086 | 2.026 | 2.354e-06 | 2.424e-05 | VATI | ENSG00000108828 | Details |
| 52. | MEGF6 | 0.026 | 0.060 | 2.335 | 3.213e-06 | 3.223e-05 | EGFL3 | ENSG00000162591 | Details |
| 53. | ABL2 | 0.030 | 0.067 | 2.247 | 3.240e-06 | 3.241e-05 | ABLL, ARG | ENSG00000143322 | Details |
| 54. | GPNMB | 0.018 | 0.047 | 2.648 | 4.358e-06 | 4.251e-05 | HGFIN, NMB, PLCA3 | ENSG00000136235 | Details |
| 55. | FAM180A | 0.011 | 0.031 | 2.717 | 0.000 | 0.001 | UNQ1940 | ENSG00000189320 | Details |
| 56. | EPG5 | 0.015 | 0.036 | 2.434 | 0.000 | 0.001 | HEEW1, KIAA1632, VICIS | ENSG00000152223 | Details |
| 57. | FAUP4 | 0.018 | 0.040 | 2.282 | 0.000 | 0.001 | PAARH | Details | |
| 58. | HPS1 | 0.020 | 0.044 | 2.241 | 0.000 | 0.001 | BLOC3S1, HPS | ENSG00000107521 | Details |
| 59. | VEGFA | 0.020 | 0.045 | 2.240 | 7.020e-05 | 0.001 | L-VEGF, MVCD1, VEGF, VPF | ENSG00000112715 | Details |
| 60. | SEZ6L2 | 0.022 | 0.047 | 2.148 | 8.737e-05 | 0.001 | BSRPA, PSK-1 | ENSG00000174938 | Details |
| 61. | NABP1 | 0.023 | 0.050 | 2.143 | 7.433e-05 | 0.001 | NABP1-OT1, OBFC2A, SOSS-B2, SSB2 | ENSG00000173559 | Details |
| 62. | SEMA3C | 0.023 | 0.048 | 2.120 | 0.000 | 0.001 | SEMAE, SemE | ENSG00000075223 | Details |
| 63. | SLC2A3 | 0.012 | 0.030 | 2.535 | 0.000 | 0.002 | GLUT3 | ENSG00000059804 | Details |
| 64. | MIR23AHG | 0.018 | 0.038 | 2.160 | 0.000 | 0.002 | Smyca | Details | |
| 65. | MEF2A | 0.019 | 0.038 | 2.046 | 0.001 | 0.004 | ADCAD1, RSRFC4, RSRFC9, mef2 | ENSG00000068305 | Details |
| 66. | SESN2 | 0.010 | 0.025 | 2.583 | 0.001 | 0.005 | HI95, SES2, SEST2 | ENSG00000130766 | Details |
| 67. | TRIM25 | 0.015 | 0.033 | 2.254 | 0.001 | 0.005 | EFP, RNF147, Z147, ZNF147 | ENSG00000121060 | Details |
| 68. | SYNGR2 | 0.021 | 0.042 | 2.014 | 0.001 | 0.005 | - | ENSG00000108639 | Details |
| 69. | GNS | 0.020 | 0.039 | 2.000 | 0.001 | 0.006 | G6S | ENSG00000135677 | Details |
| 70. | TPP1 | 0.017 | 0.037 | 2.141 | 0.001 | 0.007 | CLN2, GIG1, LPIC, SCAR7, TPP-1 | ENSG00000166340 | Details |
| 71. | EVC | 0.011 | 0.027 | 2.476 | 0.002 | 0.008 | DWF-1, EVC1, EVCL | ENSG00000072840 | Details |
| 72. | FRMD6-AS1 | 0.009 | 0.025 | 2.593 | 0.002 | 0.009 | C14orf82 | ENSG00000258537 | Details |
| 73. | FTH1P20 | 0.014 | 0.030 | 2.182 | 0.002 | 0.009 | FTHL20 | Details | |
| 74. | OSMR | 0.015 | 0.032 | 2.109 | 0.002 | 0.009 | IL-31R-beta, IL-31RB, OSMRB, OSMRbeta, PLCA1 | ENSG00000145623 | Details |
| 75. | SLC39A14 | 0.017 | 0.035 | 2.057 | 0.002 | 0.009 | HCIN, HMNDYT2, LZT-Hs4, NET34, ZIP14, cig19 | ENSG00000104635 | Details |
| 76. | MAFG | 0.015 | 0.032 | 2.073 | 0.002 | 0.010 | hMAF | ENSG00000197063 | Details |
Berezikov Lab - 2023 © ERIBA