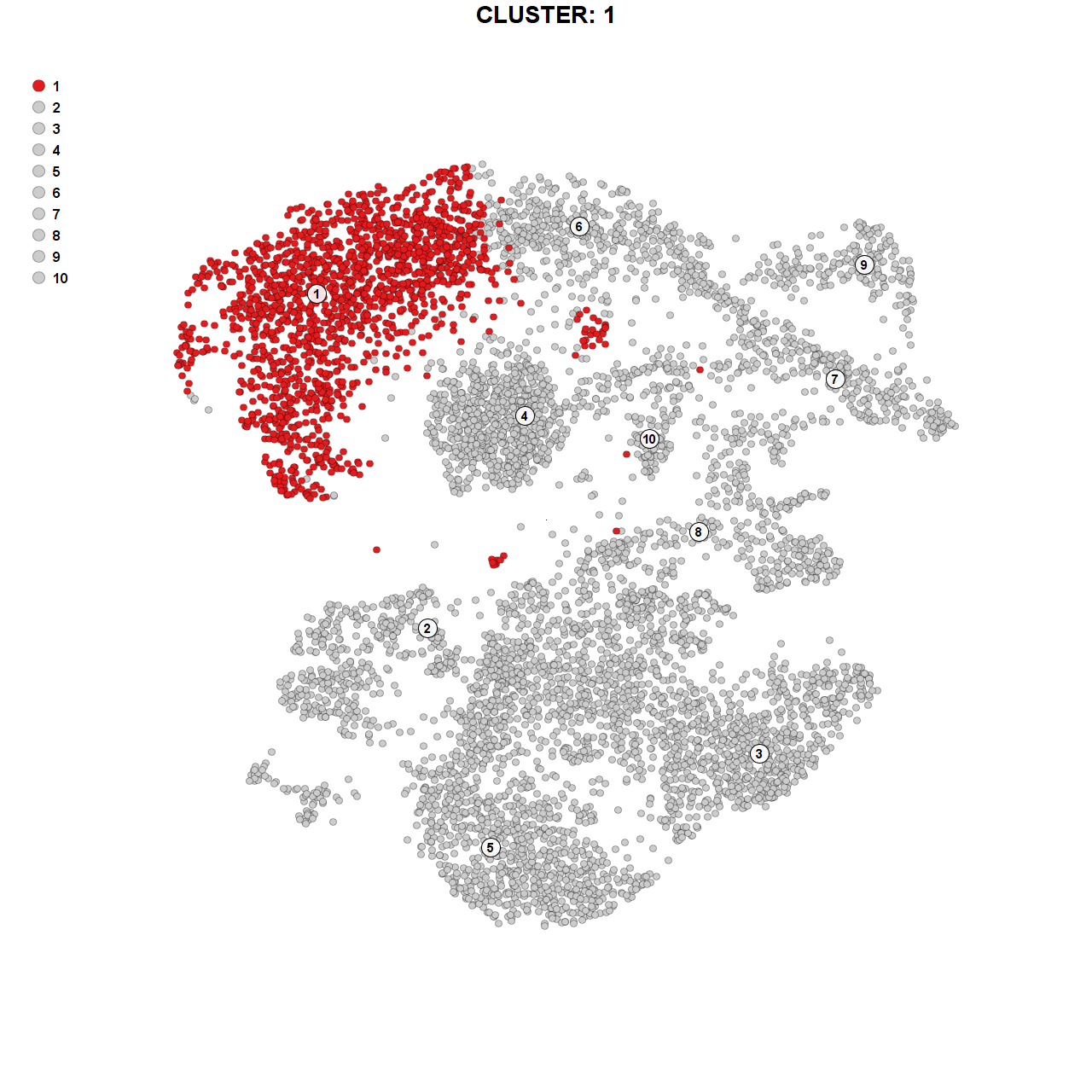
Marker genes for cluster 1
| Nr. | Gene | mean.ncl | mean.cl | fc | pv | padj | Synonyms | Ensembl | Details |
|---|---|---|---|---|---|---|---|---|---|
| 1. | PLAC8 | 0.010 | 0.025 | 2.600 | 2.944e-05 | 0.000 | C15, DGIC, PNAS-144, onzin | ENSG00000145287 | Details |
| 2. | SHROOM3 | 0.014 | 0.032 | 2.274 | 2.793e-05 | 0.000 | APXL3, MSTP013, SHRM, ShrmL | ENSG00000138771 | Details |
| 3. | DCLK2 | 0.015 | 0.033 | 2.256 | 1.618e-05 | 0.000 | CL2, CLICK-II, CLICK2, CLIK2, DCAMKL2, DCDC3, DCDC3B, DCK2 | ENSG00000170390 | Details |
| 4. | GAS1 | 0.016 | 0.035 | 2.219 | 1.076e-05 | 0.000 | - | ENSG00000180447 | Details |
| 5. | HSPB2 | 0.015 | 0.032 | 2.194 | 4.130e-05 | 0.000 | HSP27, Hs.78846, LOH11CR1K, MKBP | ENSG00000170276 | Details |
| 6. | TPD52L1 | 0.016 | 0.034 | 2.167 | 1.732e-05 | 0.000 | D53, TPD53 | ENSG00000111907 | Details |
| 7. | CALD1 | 1.301 | 2.810 | 2.160 | 0.000 | 0.000 | CDM, H-CAD, HCAD, L-CAD, LCAD, NAG22, h-CD | ENSG00000122786 | Details |
| 8. | ARMC9 | 0.020 | 0.041 | 2.017 | 1.937e-05 | 0.000 | ARM, JBTS30, KU-MEL-1, NS21 | ENSG00000135931 | Details |
| 9. | TPM1 | 0.960 | 2.030 | 2.115 | 5.894e-219 | 3.823e-216 | C15orf13, CMD1Y, CMH3, HEL-S-265, HTM-alpha, LVNC9, TMSA | ENSG00000140416 | Details |
| 10. | ID3 | 0.216 | 0.551 | 2.544 | 1.065e-86 | 2.592e-84 | HEIR-1, bHLHb25 | ENSG00000117318 | Details |
| 11. | CRYAB | 0.114 | 0.339 | 2.965 | 5.650e-69 | 9.560e-67 | CMD1II, CRYA2, CTPP2, CTRCT16, HEL-S-101, HSPB5, MFM2 | ENSG00000109846 | Details |
| 12. | RHOBTB3 | 0.128 | 0.301 | 2.342 | 7.444e-42 | 8.277e-40 | - | ENSG00000164292 | Details |
| 13. | ID1 | 0.180 | 0.368 | 2.044 | 3.545e-38 | 3.729e-36 | ID, bHLHb24 | ENSG00000125968 | Details |
| 14. | FHL1 | 0.084 | 0.209 | 2.479 | 2.312e-32 | 1.999e-30 | FCMSU, FHL-1, FHL1A, FHL1B, FLH1A, KYOT, RBMX1A, RBMX1B, SLIM, SLIM-1, SLIM1, SLIMMER, XMPMA | ENSG00000022267 | Details |
| 15. | KRT18 | 0.067 | 0.178 | 2.659 | 3.090e-31 | 2.558e-29 | CK-18, CYK18, K18 | ENSG00000111057 | Details |
| 16. | CEMIP | 0.095 | 0.222 | 2.324 | 1.349e-30 | 1.071e-28 | CCSP1, CEMIP1, HYBID, KIAA1199, TMEM2L | ENSG00000103888 | Details |
| 17. | ELN | 0.118 | 0.241 | 2.039 | 1.166e-25 | 8.254e-24 | ADCL1, SVAS, WBS, WS | ENSG00000049540 | Details |
| 18. | ALDH1A1 | 0.079 | 0.168 | 2.120 | 9.541e-20 | 4.528e-18 | ALDC, ALDH-E1, ALDH1, ALDH11, HEL-9, HEL-S-53e, HEL12, PUMB1, RALDH1 | ENSG00000165092 | Details |
| 19. | ACTA2 | 0.079 | 0.160 | 2.035 | 2.803e-17 | 1.113e-15 | ACTSA | ENSG00000107796 | Details |
| 20. | KRT7 | 0.072 | 0.150 | 2.071 | 6.146e-17 | 2.392e-15 | CK7, K2C7, K7, SCL | ENSG00000135480 | Details |
| 21. | LIMCH1 | 0.037 | 0.092 | 2.458 | 3.598e-15 | 1.262e-13 | LIMCH1A, LMO7B | ENSG00000064042 | Details |
| 22. | KRT81 | 0.019 | 0.059 | 3.110 | 4.516e-14 | 1.429e-12 | HB1, Hb-1, K81, KRTHB1, MLN137, ghHkb1, hHAKB2-1 | ENSG00000205426 | Details |
| 23. | WFDC1 | 0.013 | 0.047 | 3.636 | 1.139e-13 | 3.491e-12 | PS20 | ENSG00000103175 | Details |
| 24. | MGP | 0.031 | 0.075 | 2.462 | 8.957e-13 | 2.526e-11 | GIG36, MGLAP, NTI | ENSG00000111341 | Details |
| 25. | ACTC1 | 0.009 | 0.037 | 3.860 | 2.662e-11 | 6.515e-10 | ACTC, ASD5, CMD1R, CMH11, LVNC4 | ENSG00000159251 | Details |
| 26. | AMIGO2 | 0.014 | 0.043 | 3.152 | 1.060e-10 | 2.499e-09 | ALI1, AMIGO-2, DEGA | ENSG00000139211 | Details |
| 27. | LMOD1 | 0.032 | 0.072 | 2.255 | 2.048e-10 | 4.582e-09 | 1D, 64kD, D1, MMIHS3, SM-LMOD, SMLMOD | ENSG00000163431 | Details |
| 28. | LBH | 0.030 | 0.068 | 2.275 | 5.634e-10 | 1.218e-08 | - | ENSG00000213626 | Details |
| 29. | LMCD1 | 0.032 | 0.070 | 2.185 | 1.279e-09 | 2.593e-08 | - | ENSG00000071282 | Details |
| 30. | CLIC3 | 0.009 | 0.031 | 3.313 | 8.526e-09 | 1.603e-07 | - | ENSG00000169583 | Details |
| 31. | CSRP2 | 0.021 | 0.049 | 2.399 | 2.284e-08 | 4.155e-07 | CRP2, LMO5, SmLIM | ENSG00000175183 | Details |
| 32. | OLFML3 | 0.019 | 0.043 | 2.309 | 2.710e-07 | 4.253e-06 | HNOEL-iso, OLF44 | ENSG00000116774 | Details |
| 33. | EFHD1 | 0.017 | 0.040 | 2.303 | 2.120e-06 | 2.918e-05 | MST133, MSTP133, PP3051, SWS2 | ENSG00000115468 | Details |
| 34. | CDC42EP5 | 0.026 | 0.052 | 2.024 | 3.335e-06 | 4.460e-05 | Borg3, CEP5 | ENSG00000167617 | Details |
| 35. | NUAK1 | 0.016 | 0.031 | 2.015 | 0.000 | 0.002 | ARK5 | ENSG00000074590 | Details |
| 36. | PCDH18 | 0.011 | 0.022 | 2.045 | 0.001 | 0.009 | PCDH68L | ENSG00000189184 | Details |
Berezikov Lab - 2023 © ERIBA